Organoids as Next-Generation Models of Tumor Drug Resistance
Keywords:
Organoids, Drug resistance, 3D cultures, Microfluidic chip technologies, Preclinical models, Tumor microenvironment, Personalized medicineAbstract
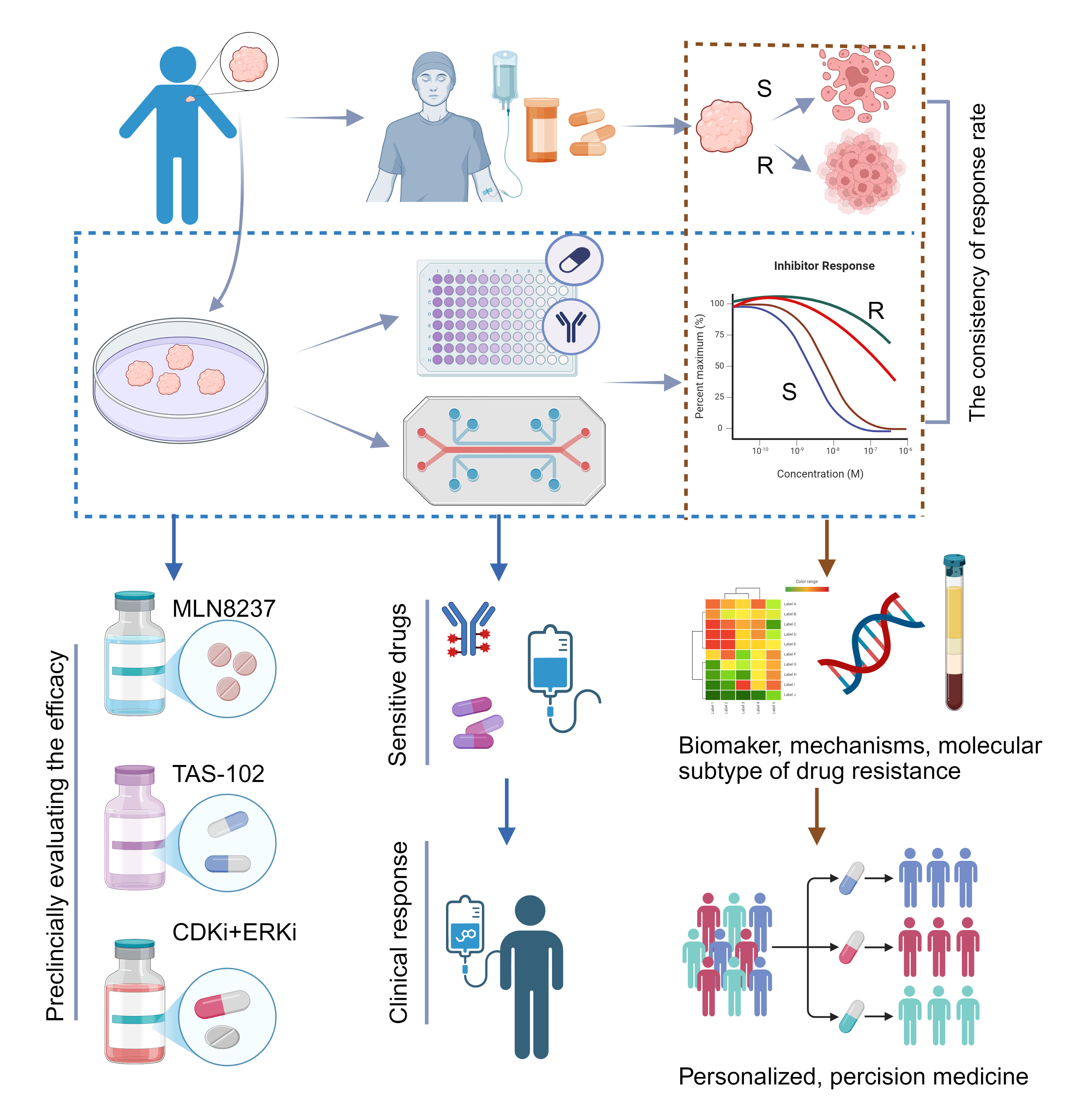
Tumor drug resistance is a major clinical challenge and critical to patient outcomes. Although extensive studies on mechanisms and drug development using cell and animal models exist, clinical results remain limited or ineffective. Recently, organoids have emerged as a powerful model that closely reproduces the structural and genetic features of tumors in vivo, making them an increasingly valuable platform for investigating resistance mechanisms and developing new therapeutic strategies. We reviewed organoid applications in elucidating tumor drug-resistance mechanisms, advancing therapies, and supporting clinical research, underscoring their critical role in overcoming treatment resistance. These mechanisms include abnormal cancer-related signaling, interactions between tumor and microenvironmental cells, DNA repair, epigenetic changes, and modifications in tumor cell membrane proteins. As an emerging preclinical model, organoids help bridge the gap between basic research and clinical practice, supporting the development of strategies to reverse drug resistance. These strategies include designing new targeted drugs, testing the efficacy of combination therapies, repurposing existing drugs, and screening personalized treatments for rare tumors. Moreover, in clinical trials, organoids can help guide therapy selection and enable preclinical evaluation of emerging therapeutic strategies. Although organoids lack the full in vivo microenvironment and an intact vascular system, advances in co-culture platforms and microfluidic chip technologies are steadily overcoming these limitations. As organoids integrate with such innovations, their role in drug-resistance research will continue to grow.
References
1. Wise JF, Lawrence MS. Huge whole-genome study of human metastatic cancers. Nature 2019;575:60–61. doi:10.1038/d41586-019-03123-0
2. Xing P, Wang S, Cao Y, Liu B, Zheng F, Guo W, et al. Treatment strategies and drug resistance mechanisms in adenocarcinoma of different organs. Drug Resist Updat. 2023;71:101002. doi:10.1016/j.drup.2023.101002
3. Nussinov R, Tsai CJ, Jang H. Anticancer drug resistance: an update and perspective. Drug Resist Updat. 2021;59:100796. doi:10.1016/j.drup.2021.100796
4. Peng Z, Lv X, Sun H, Zhao L, Huang S. 3D tumor cultures for drug resistance and screening development in clinical applications. Mol Cancer. 2025;24:93. doi:10.1186/s12943-025-02281-2
5. Chai C, Ji P, Xu H, Tang H, Wang Z, Zhang H, et al. Targeting cancer drug resistance utilizing organoid technology. Biomed Pharmacother. 2023;158:114098. doi:10.1016/j.biopha.2022.114098
6. Rahmanian M, Seyfoori A, Ghasemi M, Shamsi M, Rezaei Kolahchi A, Pezeshgi Modarres H, et al. In-vitro tumor microenvironment models containing physical and biological barriers for modelling multidrug resistance mechanisms and multidrug delivery strategies. J Control Release. 2021;334:164–177. doi:10.1016/j.jconrel.2021.04.024
7. Wang Q, Yuan F, Zuo X, Li M. Breakthroughs and challenges of organoid models for assessing cancer immunotherapy: a cutting-edge tool for advancing personalised treatments. Cell Death Discov. 2025;11:222. doi:10.1038/s41420-025-02505-w
8. Singh D, Thakur A, Rakesh, Kumar A. Advancements in organoid-based drug discovery: revolutionizing precision medicine and Pharmacology. Drug Dev Res. 2025;86:e70121. doi:10.1002/ddr.70121
9. Wang Y, Sun X, Lu B, Zhang D, Yin Y, Liu S, et al. Current applications, future Perspectives and challenges of Organoid technology in oral cancer research. Eur J Pharmacol. 2025;993:177368. doi:10.1016/j.ejphar.2025.177368
10. Li Z, Li K, Zhang C, Zhao Y, Guo Y, He J, et al. Bioprinted organoids: an innovative engine in biomedicine. Adv Sci (Weinh). 2025;12(33):e07317. doi:10.1002/advs.202507317
11. El Harane S, Nazari B, El Harane N, Locatelli M, Zidi B, Durual S, et al. Generation of individualized, standardized, and electrically synchronized human midbrain organoids. Cells. 2025;14(15):1211. doi:10.3390/cells14151211
12. Wu J, Liu T, Zhang X, Qu C, Wu J, Xu S, et al. Progress in the application of organoids for exploring the relationship between macrophages and various lung diseases. Biofabrication. 2025;17(3). doi:10.1088/1758-5090/adde15
13. Ding Z, Chang X, Qu X, Hua K, Qiu J. Gynecological malignancy organoids: A game changer for personalized medicine. Biochim Biophys Acta Rev Cancer. 2025;1880:189405. doi:10.1016/j.bbcan.2025.189405
14. Li Q, Xiao Y, Han L, Luo W, Dai W, Fang H, et al. Microbiome dysbiosis, neutrophil recruitment and mesenchymal transition of mesothelial cells promotes peritoneal metastasis of colorectal cancer. Nat Cancer. 2025;6(3):493-510. doi:10.1038/s43018-025-00910-9
15. Boilève A, Cartry J, Goudarzi N, Bedja S, Mathieu JRR, Bani MA, et al. Organoids for functional precision medicine in advanced pancreatic cancer. Gastroenterology. 2024;167(5):961-976.e13. doi:10.1053/j.gastro.2024.05.032
16. Sun X, Cai W, Li H, Gao C, Ma X, Guo Y, et al. Endothelial-like cancer-associated fibroblasts facilitate pancreatic cancer metastasis via vasculogenic mimicry and paracrine signalling. Gut. 2025;74(9):1437-1451. doi:10.1136/gutjnl-2024-333638
17. Yang R, Wang S, Li Z, Yin C, Huang W, Huang W. Patient-derived organoid co-culture systems as next-generation models for bladder cancer stem cell research. Cancer Lett. 2025;625:217793. doi:10.1016/j.canlet.2025.217793
18. Li Y, Liu J, Xu S, Wang J. 3D bioprinting: an important tool for tumor microenvironment research. Int J Nanomedicine. 2023;18:8039-8057. doi:10.2147/IJN.S435845
19. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19(2):65-81. doi:10.1038/s41568-018-0104-6
20. Mi W, van Tienderen GS, Shi S, Broeders A, Monfils K, Roest HP, et al. Apoptosis regulators of the Bcl-2 family play a key role in chemoresistance of cholangiocarcinoma organoids. Int J Cancer. 2025;157(8):1694-1708. doi:10.1002/ijc.35483
21. Álvarez-Varela A, Novellasdemunt L, Barriga FM, Hernando-Momblona X, Cañellas-Socias A, Cano-Crespo S, et al. Mex3a marks drug-tolerant persister colorectal cancer cells that mediate relapse after chemotherapy. Nat Cancer. 2022;3(9):1052-1070. doi:10.1038/s43018-022-00402-0
22. Yang R, Kwan W, Du Y, Yan R, Zang L, Li C, et al. Drug-induced senescence by aurora kinase inhibitors attenuates innate immune response of macrophages on gastric cancer organoids. Cancer Lett. 2024;598:217106. doi:10.1016/j.canlet.2024.217106
23. Würth R, Donato E, Michel LL, Saini M, Becker L, Cheytan T, et al. Circulating tumor cell plasticity determines breast cancer therapy resistance via neuregulin 1-HER3 signaling. Nat Cancer. 2025;6(1):67-85. doi:10.1038/s43018-024-00882-2
24. Tong X, Patel AS, Kim E, Li H, Chen Y, Li S, et al. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. Cancer Cell. 2024;42(3):413-428.e7. doi:10.1016/j.ccell.2024.01.012
25. Sase M, Sato T, Sato H, Miya F, Zhang S, Haeno H, et al. Comparative analysis of tongue cancer organoids among patients identifies the heritable nature of minimal residual disease. Dev Cell. 2025;60(3):396-413.e6. doi:10.1016/j.devcel.2024.10.007
26. Mosquera MJ, Kim S, Bareja R, Fang Z, Cai S, Pan H, et al. Extracellular matrix in synthetic hydrogel-based prostate cancer organoids regulate therapeutic response to EZH2 and DRD2 inhibitors. Adv Mater. 2022;34(2):e2100096. doi:10.1002/adma.202100096
27. Yang D, Zhang X, Hu Z, Sun Q, Fu H, Yao J, et al. Organoid-based single cell sequencing revealed the lineage evolution during docetaxel treatment in gastric cancer. Cancer Lett. 2025;619:217617. doi:10.1016/j.canlet.2025.217617
28. Mancini C, Lori G, Mattei G, Iozzo M, Desideri D, Cianchi F, et al. PHGDH drives 5-FU chemoresistance in colorectal cancer through the Hedgehog signaling. J Exp Clin Cancer Res. 2025;44(1):198. doi:10.1186/s13046-025-03447-y
29. Xu Q, Jiang Z, Pan Y, Li S, Cao Z, Hua S, et al. Cucurbitacin B stimulates PD-1 immunotherapy response in malignant breast cancer by covalent targeting MTCH2. 2025;145:157017. doi:10.1016/j.phymed.2025.157017
30. Muniyan S, Vengoji R, Nimmakayala RK, Seshacharyulu P, Perumalsamy B, Alsafwani ZW, et al. PAF1-mediated transcriptional reprogramming confers docetaxel resistance in advanced prostate cancer. Cancer Lett. 2025;609:217355. doi:10.1016/j.canlet.2024.217355
31. Fei Y, Cao D, Li Y, Wang Z, Dong R, Zhu M, et al. Circ_0008315 promotes tumorigenesis and cisplatin resistance and acts as a nanotherapeutic target in gastric cancer. J Nanobiotechnology. 2024;22:519. doi:10.1186/s12951-024-02760-6
32. Ren T, Qiu J, Chen F, Jiang Q, Liu Q, Wu T, et al. Targeting glutamine metabolism transporter SLC25A22 enhances CD8+ T-cell function and anti-PD-1 therapy efficacy in cervical squamous cell carcinoma: integrated metabolomics, transcriptomics and T-cell-incorporated tumor organoid studies. Adv Sci (Weinh). 2025;12:e02225. doi:10.1002/advs.202502225
33. Yao N, Jing N, Lin J, Niu W, Yan W, Yuan H, et al. Patient-derived tumor organoids for cancer immunotherapy: culture techniques and clinical application. Invest New Drugs. 2025;43:394–404. doi:10.1007/s10637-025-01523-w
34. Goto H, Nishioka Y, et al. Goto H, Nishioka Y. Fibrocytes: a novel stromal cells to regulate resistance to anti-angiogenic therapy and cancer progression. Int J Mol Sci. 2017;19:98. doi:10.3390/ijms19010098
35. Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi:10.1038/s41591-018-0096-5
36. Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, et al. Cancer-associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell Mol Gastroenterol Hepatol. 2021;11:407–431. doi:10.1016/j.jcmgh.2020.09.003
37. Guo Y, Li Q, Ye Q, Jin Y, Yu Y, Zhang X, et al. Construction and drug screening of co-culture system using extrahepatic cholangiocarcinoma organoids and tumor-associated macrophages. Heliyon. 2024;10:e36377. doi:10.1016/j.heliyon.2024.e36377
38. Kang SH, Oh SY, Lee KY, Lee HJ, Kim MS, Kwon TG, et al. Differential effect of cancer-associated fibroblast-derived extracellular vesicles on cisplatin resistance in oral squamous cell carcinoma via miR-876-3p. Theranostics. 2024;14:460–479. doi:10.7150/thno.87329
39. Farin HF, Mosa MH, Ndreshkjana B, Grebbin BM, Ritter B, Menche C, et al. Colorectal cancer organoid-stroma biobank allows subtype-specific assessment of individualized therapy responses. Cancer Discov. 2023;13:2192–2211. doi:10.1158/2159-8290.CD-23-0050
40. Ma Y, Xue F, Pei Z, Zhao Y. Constructing a co-culture model of cancer-associated fibroblasts and ovarian cancer organoids and studying mechanisms of drug resistance. Exp Cell Res. 2025;450:114656. doi:10.1016/j.yexcr.2025.114656
41. Ma B, Ma C, Long X, Jiang L. Stromal reprogramming in urachal cancer: fibroblast activation protein and collagen remodeling drive immune-suppressive niches and immunotherapy resistance. Int Immunopharmacol. 2025;163:115204. doi:10.1016/j.intimp.2025.115204
42. Ryu KB, Seo JA, Lee K, Choi J, Yoo G, Ha JH, et al. Drug-resistance biomarkers in patient-derived colorectal cancer organoid and fibroblast co-culture system. Curr Issues Mol Biol. 2024;46:5794–5811. doi:10.3390/cimb46060346
43. Roh HS, Kim DE, Kim G, Kim J, Fan D, Kim HS, et al. Establishment and long-term expansion of adult hepatobiliary organoids co-cultured with liver endothelial cells. Heliyon. 2024;10:e36120. doi:10.1016/j.heliyon.2024.e36120
44. Lin CN, Liang YL, Tsai HF, Wu PY, Huang LY, Lin YH, et al. Adipocyte pyroptosis occurs in omental tumor microenvironment and is associated with chemoresistance of ovarian cancer. J Biomed Sci. 2024;31:62. doi:10.1186/s12929-024-01051-4
45. Jiang S, Deng T, Cheng H, Liu W, Shi D, Yuan J, et al. Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J Exp Clin Cancer Res. 2023;42:199. doi:10.1186/s13046-023-02756-4
46. Shi F, Li GJ, Liu Y, Zhou HM, Zhang Y, Wei SY, et al. USP19 deficiency enhances T-cell-mediated antitumor immunity by promoting PD-L1 degradation in colorectal cancer. Pharmacol Res. 2025;214:107668. doi:10.1016/j.phrs.2025.107668
47. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598.e12. doi:10.1016/j.cell.2018.07.009
48. Subtil B, Cambi A, Tauriello DVF, de Vries IJM. The therapeutic potential of tackling tumor-induced dendritic cell dysfunction in colorectal cancer. Front Immunol. 2021;12:724883. doi:10.3389/fimmu.2021.724883
49. LeSavage BL, Zhang D, Huerta-López C, Gilchrist AE, Krajina BA, Karlsson K, et al. Engineered matrices reveal stiffness-mediated chemoresistance in patient-derived pancreatic cancer organoids. Nat Mater. 2024;23:1138–1149. doi:10.1038/s41563-024-01908-x
50. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e16. doi:10.1016/j.cell.2018.11.021
51. Zhang B, Ohuchida K, Tsutsumi C, Shimada Y, Mochida Y, Oyama K, et al. Dynamic glycolytic reprogramming effects on dendritic cells in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2024;43:271. doi:10.1186/s13046-024-03192-8
52. Zhang J, Tavakoli H, Ma L, Li X, Han L, Li X. Immunotherapy discovery on tumor organoid-on-a-chip platforms that recapitulate the tumor microenvironment. Adv Drug Deliv Rev. 2022;187:114365. doi:10.1016/j.addr.2022.114365
53. Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi:10.1158/2159-8290.CD-17-0833
54. Sethakorn N, Heninger E, Breneman MT, Recchia E, Ding AB, Jarrard DF, et al. Integrated analysis of the tumor microenvironment using a reconfigurable microfluidic cell culture platform. FASEB J. 2022;36:e22540. doi:10.1096/fj.202200684RR
55. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi:10.1038/nbt.1685
56. Gunjan A, Singh RK. Epigenetic therapy: targeting histones and their modifications in human disease. Future Med Chem. 2010;2:543–548. doi:10.4155/fmc.10.18
57. Orsolic I, Carrier A, Esteller M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet. 2023;39:74–88. doi:10.1016/j.tig.2022.10.004
58. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi:10.1016/j.cell.2017.05.045
59. Wang D, Zhang Y, Li Q, Zhang A, Xu J, Li Y, et al. N6 methyladenosine (m6A) in cancer therapeutic resistance: potential mechanisms and clinical implications. Biomed Pharmacother. 2023;167:115477. doi:10.1016/j.biopha.2023.115477
60. Kraiczy J, Nayak KM, Howell KJ, Ross A, Forbester J, Salvestrini C, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi:10.1136/gutjnl-2017-314817
61. Ghorbaninejad M, Asadzadeh Aghdaei H, Baharvand H, Meyfour A. Intestinal organoids: a versatile platform for modeling gastrointestinal diseases and monitoring epigenetic alterations. Life Sci. 2023;319:121506. doi:10.1016/j.lfs.2023.121506
62. Nguyen TBN, Gevers S, Kok RNU, Burgering LM, Neikes H, Akkerman N, et al. Lactate controls cancer stemness and plasticity through epigenetic regulation. Cell Metab. 2025;37:903–919.e10. doi:10.1016/j.cmet.2025.01.002
63. Silva Almeida C, Ewart MA, Wilde C. 3D gastrointestinal models and organoids to study metabolism in human colon cancer. Semin Cell Dev Biol. 2020;98:98–104. doi:10.1016/j.semcdb.2019.05.019
64. Yu M, Ni M, Xu F, Liu C, Chen L, Li J, et al. NSUN6 mediated 5 methylcytosine modification of NDRG1 mRNA promotes radioresistance in cervical cancer. Mol Cancer. 2024;23:139. doi:10.1186/s12943-024-02055-2
65. Zhang X, Su T, Wu Y, Cai Y, Wang L, Liang C, et al. N6 methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. 2024;84:827–840. doi:10.1158/0008-5472.CAN-23-1916
66. Xie R, Cheng L, Huang M, Huang L, Chen Z, Zhang Q, et al. NAT10 drives cisplatin chemoresistance by enhancing ac4C associated DNA repair in bladder cancer. Cancer Res. 2023;83:1666–1683. doi:10.1158/0008-5472.CAN-22-2233
67. Xu X, Wang Q, Guo K, Xu J, Lu Y, Chen H, et al. CD47 blockade reverses resistance to HDAC inhibitor by liberating anti tumor capacity of macrophages. J Exp Clin Cancer Res. 2025;44:67. doi:10.1186/s13046-025-03335-5
68. Zhang M, Zhong A, Liu H, Zhao L, Wang Y, Lu Z, et al. EZH2 loss promotes gastric squamous cell carcinoma. Nat Commun. 2025;16:6032. doi:10.1038/s41467-025-61024-5
69. Wang L, Wang X, Zhu X, Zhong L, Jiang Q, Wang Y, et al. Drug resistance in ovarian cancer: from mechanism to clinical trial. Mol Cancer. 2024;23:66. doi:10.1186/s12943-024-01967-3
70. Ikeuchi H, Matsuno Y, Kusumoto Matsuo R, Kojima S, Ueno T, Ikegami M, et al. GLI1 confers resistance to PARP inhibitors by activating the DNA damage repair pathway. Oncogene. 2024;43:3037–3048. doi:10.1038/s41388-024-03105-1
71. Ubhi T, Zaslaver O, Quaile AT, Plenker D, Cao P, Pham NA, Békési A, et al. Cytidine deaminases APOBEC3C and APOBEC3D promote DNA replication stress resistance in pancreatic cancer cells. Nat Cancer. 2024;5:895–915. doi:10.1038/s43018-024-00742-z
72. Stoof J, Harrold E, Mariottino S, Lowery MA, Walsh N. DNA damage repair deficiency in pancreatic ductal adenocarcinoma: preclinical models and clinical perspectives. Front Cell Dev Biol. 2021;9:749490. doi:10.3389/fcell.2021.749490
73. Ribeiro CF, Rodrigues S, Bastos DC, Fanelli GN, Pakula H, Foiani M, et al. Blocking lipid synthesis induces DNA damage in prostate cancer and increases cell death caused by PARP inhibition. Sci Signal. 2024;17:eadh1922. doi:10.1126/scisignal.adh1922
74. Qu S, Qi S, Zhang H, Li Z, Wang K, Zhu T, et al. Albumin-bound paclitaxel augments temozolomide treatment sensitivity of glioblastoma cells by disrupting DNA damage repair and promoting ferroptosis. J Exp Clin Cancer Res. 2023;42:285. doi:10.1186/s13046-023-02843-6
75. Chen H, Li Y, Li H, Chen X, Fu H, Mao D, et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature. 2024;631:663–669. doi:10.1038/s41586-024-07620-9
76. Chen Y, Wu J, Zhai L, Zhang T, Yin H, Gao H, et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell. 2024;187:294–311.e21. doi:10.1016/j.cell.2023.11.022
77. Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, et al. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. doi:10.1016/j.molcel.2016.06.020
78. Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology mediated end joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–237. doi:10.1038/nsmb.2961
79. Zatreanu D, Robinson HMR, Alkhatib O, Boursier M, Finch H, Geo L, et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat Commun. 2021;12:3636. doi:10.1038/s41467-021-23463-8
80. Blukacz L, Nuciforo S, Fucile G, Trulsson F, Duthaler U, Wieland S, et al. Inhibition of the transmembrane transporter ABCB1 overcomes resistance to doxorubicin in patient-derived organoid models of HCC. Hepatol Commun. 2024;8:e0000000000000437. doi:10.1097/HC9.0000000000000437
81. Nimmakayala RK, Leon F, Rachagani S, Rauth S, Nallasamy P, Marimuthu S, et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene. 2021;40:215–231. doi:10.1038/s41388-020-01518-2
82. Chau CH, Steeg PS, Figg WD. Antibody drug conjugates for cancer. Lancet. 2019;394:793–804. doi:10.1016/S0140-6736(19)31774-X
83. Benelli R, Costa D, Barboro P, Poggi A, Matis S, Zocchi MR, et al. Targeting colorectal cancer organoids with zoledronic acid conjugated to the anti-EGFR antibody cetuximab. J Immunother Cancer. 2022;10:e005660. doi:10.1136/jitc-2022-005660
84. Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389-1400. doi:10.1038/nm.3388
85. Cortés J, Kim SB, Chung WP, Im SA, Park YH, Hegg R, et al. Trastuzumab deruxtecan versus trastuzumab emtansine for breast cancer. N Engl J Med. 2022;386:1143–1154. doi:10.1056/NEJMoa2115022
86. Díaz Rodríguez E, Gandullo Sánchez L, Ocaña A, Pandiella A. Novel ADCs and strategies to overcome resistance to anti-HER2 ADCs. Cancers (Basel). 2021;14:154. doi:10.3390/cancers14010154
87. Wolf DM, Yau C, Wulfkuhle J, Brown Swigart L, Gallagher RI, Lee PRE, et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: predictive biomarkers across 10 cancer therapies. Cancer Cell. 2022;40:609–623.e6. doi:10.1016/j.ccell.2022.05.005
88. Chen YF, Zhang QH, Zhang ZW, Zhou YJ, Liu CC, Shao ZM, et al. SNX10 deficiency impairs sensitivity to anti-HER2 antibody–drug conjugates via altering HER2 trafficking in HER2-positive breast cancer. Proc Natl Acad Sci U S A. 2025;122:e2417586122. doi:10.1073/pnas.2417586122
89. Rathje F, Klingler S, Aberger F. Organoids for modeling (colorectal) cancer in a dish. Cancers (Basel). 2022;14:5416. doi:10.3390/cancers14215416
90. Tardito S, Matis S, Zocchi MR, Benelli R, Poggi A. Epidermal growth factor receptor targeting in colorectal carcinoma: antibodies and patient derived organoids as a smart model to study therapy resistance. Int J Mol Sci. 2024;25:7131. doi:10.3390/ijms25137131
91. Liu T, Wang H, Chen Y, Wan Z, Du Z, Shen H, et al. SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation. J Exp Clin Cancer Res. 2023;42(1):234. doi:10.1186/s13046-023-02789-9
92. Elurbide J, Colyn L, Latasa MU, Uriarte I, Mariani S, Lopez Pascual A, et al. Identification of PRMT5 as a therapeutic target in cholangiocarcinoma. Gut. 2024;74(1):116-127. doi:10.1136/gutjnl-2024-332998
93. Zhang T, Febres Aldana C, Liu Z, Dix JM, Cheng R, Dematteo RG, et al. HER2 antibody-drug conjugates are active against desmoplastic small round cell tumor. Clin Cancer Res. 2024;30:4701-4713. doi:10.1158/1078-0432.CCR-24-1835
94. Weng W, Meng T, Zhao Q, Shen Y, Fu G, Shi J, et al. Antibody-Exatecan conjugates with a novel self-immolative moiety overcome resistance in colon and lung cancer. Cancer Discov. 2023;13:950-973. doi:10.1158/2159-8290.CD-22-1368
95. Hong X, Chen X, Wang H, Xu Q, Xiao K, Zhang Y, et al. A HER2-targeted antibody-drug conjugate, RC48-ADC, exerted promising antitumor efficacy and safety with intravesical instillation in preclinical models of bladder cancer. Adv Sci (Weinh). 2023;10:e2302377. doi:10.1002/advs.202302377
96. Guo C, Figueiredo I, Gurel B, Neeb A, Seed G, Crespo M, et al. B7-H3 as a therapeutic target in advanced prostate cancer. Eur Urol. 2023;83:e168-e169. doi:10.1016/j.eururo.2022.09.004
97. Miranda Furtado CL, Dos Santos Luciano MC, Silva Da Santos R, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019;14:1164-1176. doi:10.1080/15592294.2019.1640546
98. Parizadeh SM, Jafarzadeh Esfehani R, Ghandehari M, Seifi S, Parizadeh SMR, Moetamani-Ahmadi M, et al. Epigenetic drug therapy in the treatment of colorectal cancer. Curr Pharm Des. 2018;24:2701-2709. doi:10.2174/1381612824666180730151904
99. Morgan AG, Griffin MF, Longaker MT, Norton JA. Precision medicine: IL-1RA and pancreatic cancer organoids. Biology (Basel). 2025;14:60604. doi:10.3390/biology14060604
100. Liu C, Li J, Xu F, Chen L, Ni M, Wu J, et al. PARP1-DOT1L transcription axis drives acquired resistance to PARP inhibitor in ovarian cancer. Mol Cancer. 2024;23:111. doi:10.1186/s12943-024-02025-8
101. Wang F, Yu X, Qian J, Cao Y, Dong S, Zhan S, et al. A novel SIK2 inhibitor SIC-19 exhibits synthetic lethality with PARP inhibitors in ovarian cancer. Drug Resist Updat. 2024;74:101077. doi:10.1016/j.drup.2024.101077
102. Spender LC, Watt DM, Baxter MA, Shuttleworth MK, Walker K, Savage AR, et al. AKT/mTOR as a targetable hub to overcome multimodal resistance to EGFR inhibitors in oesophageal squamous cell carcinoma. Br J Cancer. 2025;133(5):709-722. doi:10.1038/s41416-025-03093-3
103. Zhou Z, Van der Jeught K, Li Y, Sharma S, Yu T, Moulana I, et al. A T cell-engaging tumor organoid platform for pancreatic cancer immunotherapy. Adv Sci (Weinh). 2023;10:e2300548. doi:10.1002/advs.202300548
104. Xia Y, Sun M, Huang H, Jin WL. Drug repurposing for cancer therapy. Signal Transduct Target Ther. 2024;9(1):92. doi:10.1038/s41392-024-01808-1
105. Sada N, Lee S, Katsu T, Otsuki T, Inoue T. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science. 2015;347:1362-1367. doi:10.1126/science.aaa1299
106. Frampton JE. Stiripentol: a review in Dravet syndrome. Drugs. 2019;79:1785-1796. doi:10.1007/s40265-019-01204-y
107. Xie CK, Liao CY, Lin HY, Wu YD, Lu FC, Huang XX, et al. Sulindac (K-80003) with nab-paclitaxel and gemcitabine overcomes drug-resistant pancreatic cancer. Mol Cancer. 2024;23:215. doi:10.1186/s12943-024-02128-2
108. Suvilesh KN, Manjunath Y, Nussbaum YI, Gadelkarim M, Raju M, Srivastava A, et al. Targeting AKR1B10 by drug repurposing with epalrestat overcomes chemoresistance in non–small cell lung cancer patient-derived tumor organoids. Clin Cancer Res. 2024;30(17):3855-3867. doi:10.1158/1078-0432.CCR-23-3980
109. Yehya A, Ghamlouche F, Karami R, Hachem S, Salhab Z, Liu YN, et al. Repurposing piroxicam enhances the antineoplastic effects of docetaxel and enzalutamide in prostate cancer cells using 2D and 3D in vitro culture models. Front Cell Dev Biol. 2025;13:1551010. doi:10.3389/fcell.2025.1551010
110. Sun L, Kang X, Ju H, Wang C, Yang G, Wang R, et al. A human mucosal melanoma organoid platform for modeling tumor heterogeneity and exploring immunotherapy combination options. Sci Adv. 2023;9(43):eadg6686. doi:10.1126/sciadv.adg6686
111. Al Shihabi A, Tebon PJ, Nguyen HTL, Chantharasamee J, Sartini S, Davarifar A, et al. The landscape of drug sensitivity and resistance in sarcoma. Cell Stem Cell. 2024;31:1524-1542.e1524. doi:10.1016/j.stem.2024.08.010
112. Kim SY, de Weert TAE, Vermeulen M, Ringnalda F, Kester L, Zsiros J, et al. Organoid drug profiling identifies methotrexate as a therapy for SCCOHT, a rare pediatric cancer. Sci Adv. 2025;11:eadq1724. doi:10.1126/sciadv.adq1724
113. Verstegen MMA, Coppes RP, Beghin A, De Coppi P, Gerli MFM, de Graeff N, et al. Clinical applications of human organoids. Nat Med. 2025;31:409-421. doi:10.1038/s41591-024-03489-3
114. Zhao Y, Li S, Zhu L, Huang M, Xie Y, Song X, et al. Personalized drug screening using patient-derived organoid and its clinical relevance in gastric cancer. Cell Rep Med. 2024;5:101627. doi:10.1016/j.xcrm.2024.101627
115. Jensen LH, Rogatto SR, Lindebjerg J, Havelund B, Abildgaard C, do Canto LM, et al. Precision medicine applied to metastatic colorectal cancer using tumor-derived organoids and in-vitro sensitivity testing: a phase 2, single-center, open-label, and non-comparative study. J Exp Clin Cancer Res. 2023;42:115. doi:10.1186/s13046-023-02683-4
116. Ma YS, Yang XL, Xin R, Wu TM, Shi Y, Zhang DD, et al. The power and the promise of organoid models for cancer precision medicine with next-generation functional diagnostics and pharmaceutical exploitation. Transl Oncol.2021;14:101126. doi:10.1016/j.tranon.2021.101126
117. van de Haar J, Ma X, Ooft SN, van der Helm PW, Hoes LR, Mainardi S, et al. Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat Med. 2023;29:605-614. doi:10.1038/s41591-023-02240-8
118. Hedayat S, Cascione L, Cunningham D, Schirripa M, Lampis A, Hahne JC, et al. Circulating microRNA analysis in a prospective co-clinical trial identifies MIR652-3p as a response biomarker and driver of regorafenib resistance mechanisms in colorectal cancer. Clin Cancer Res. 2024;30:2140-2159. doi:10.1158/1078-0432.ccr-23-2748
119. Mayer RJ, Van Cutsem E, Falcone A, Yoshino T, Garcia-Carbonero R, Mizunuma N, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372:1909-1919. doi:10.1056/NEJMoa1414325
120. Van Cutsem E, Mayer RJ, Laurent S, Winkler R, Grávalos C, Benavides M, et al. The subgroups of the phase III RECOURSE trial of trifluridine/tipiracil (TAS-102) versus placebo with best supportive care in patients with metastatic colorectal cancer. Eur J Cancer. 2018;90:63-72. doi:10.1016/j.ejca.2017.10.009
121. Beltran H, Oromendia C, Danila DC, Montgomery B, Hoimes C, Szmulewitz RZ, et al. A phase II trial of the Aurora kinase A inhibitor alisertib for patients with castration-resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin Cancer Res. 2019;25:43-51. doi:10.1158/1078-0432.ccr-18-1912
122. Goodwin CM, Waters AM, Klomp JE, Javaid S, Bryant KL, Stalnecker C, et al. Combination therapies with CDK4/6 inhibitors to treat KRAS-mutant pancreatic cancer. Cancer Res. 2023;83:141-157. doi:10.1158/0008-5472.can-22-0391
123. Xu H, Jiao D, Liu A, Wu K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol. 2022;15:58. doi:10.1186/s13045-022-01278-4
124. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373-386.e310. doi:10.1016/j.cell.2017.11.010
125. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol.2020;21:571-584. doi:10.1038/s41580-020-0259-3
126. Koh V, Chakrabarti J, Torvund M, Steele N, Hawkins JA, Ito Y, et al. Hedgehog transcriptional effector GLI mediates mTOR-induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021;518:59-71. doi:10.1016/j.canlet.2021.06.007
127. Holokai L, Chakrabarti J, Lundy J, Croagh D, Adhikary P, Richards SS, et al. Murine- and human-derived autologous organoid/immune cell co-cultures as pre-clinical models of pancreatic ductal adenocarcinoma. Cancers (Basel). 2020;12:3816. doi:10.3390/cancers12123816
128. Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10:1129-1139. doi:10.1158/2159-8290.cd-20-0187
129. Wang K, Yuen ST, Xu J, Lee SP, Yan HHN, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. doi:10.1038/ng.2983
130. Zhao Y, Li ZX, Zhu YJ, Fu J, Zhao XF, Zhang YN, et al. Single-cell transcriptome analysis uncovers intratumoral heterogeneity and underlying mechanisms for drug resistance in hepatobiliary tumor organoids. Adv Sci (Weinh). 2021;8:e2003897. doi:10.1002/advs.202003897
131. Wang G, Xu Y, Wang Q, Chai Y, Sun X, Yang F, et al. Rare and undiagnosed diseases: from disease-causing gene identification to mechanism elucidation. Fundam Res. 2022;2:918-928. doi:10.1016/j.fmre.2022.09.002
132. Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515-528.e517. doi:10.1016/j.cell.2018.03.017
133. Maru Y, Tanaka N, Itami M, Hippo Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol Oncol. 2019;154:189-198. doi:10.1016/j.ygyno.2019.05.005
134. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920-926. doi:10.1126/science.aao2774
135. Gronholm M, Feodoroff M, Antignani G, Martins B, Hamdan F, Cerullo V. Patient-derived organoids for precision cancer immunotherapy. Cancer Res. 2021;81:3149-3155. doi:10.1158/0008-5472.can-20-4026
136. Zhou X, Qu M, Tebon P, Jiang X, Wang C, Xue Y, et al. Screening cancer immunotherapy: when engineering approaches meet artificial intelligence. Adv Sci (Weinh). 2020;7:2001447. doi:10.1002/advs.202001447
137. Huang Y, Huang Z, Tang Z, Chen Y, Huang M, Liu H, et al. Research progress, challenges, and breakthroughs of organoids as disease models. Front Cell Dev Biol. 2021;9:740574. doi:10.3389/fcell.2021.740574
138. Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653-658. doi:10.1016/j.stem.2013.11.002
139. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407-418. doi:10.1038/s41568-018-0007-6
140. De Sousa E Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676-680. doi:10.1038/nature21713
141. Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8:745-760. doi:10.15252/emmm.201606184
142. Michels BE, Mosa MH, Streibl BI, Zhan T, Menche C, Abou El Ardat K, et al. Pooled in vitro and in vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell. 2020;26:782-792.e787. doi:10.1016/j.stem.2020.04.003
143. Yang J, Huang S, Cheng S, Jin Y, Zhang N, Wang Y. Application of ovarian cancer organoids in precision medicine: key challenges and current opportunities. Front Cell Dev Biol. 2021;9:701429. doi:10.3389/fcell.2021.701429
144. Liang J, Wei J, Cao J, Qian J, Gao R, Li X, et al. In-organoid single-cell CRISPR screening reveals determinants of hepatocyte differentiation and maturation. Genome Biol. 2023;24:251. doi:10.1186/s13059-023-03084-8
145. Guo D, Yao B, Shao WW, Zuo JC, Chang ZH, Shi JX, et al. The critical role of YAP/BMP/ID1 axis on simulated microgravity-induced neural tube defects in human brain organoids. Adv Sci (Weinh). 2025;12:e2410188. doi:10.1002/advs.202410188
146. Bai L, Wu Y, Li G, Zhang W, Zhang H, Su J. AI-enabled organoids: construction, analysis, and application. Bioact Mater. 2024;31:525-548. doi:10.1016/j.bioactmat.2023.09.005
147. Man Y, Liu Y, Chen Q, Zhang Z, Li M, Xu L, et al. Organoids-on-a-chip for personalized precision medicine. Adv Healthc Mater. 2024;13:e2401843. doi:10.1002/adhm.202401843
148. Wahida A, Buschhorn L, Frohling S, Jost PJ, Schneeweiss A, Lichter P, et al. The coming decade in precision oncology: six riddles. Nat Rev Cancer. 2023;23:43-54. doi:10.1038/s41568-022-00529-3
149. Johansson A, Andreassen OA, Brunak S, Franks PW, Hedman H, Loos RJF, et al. Precision medicine in complex diseases - molecular subgrouping for improved prediction and treatment stratification. J Intern Med. 2023;294:378-396. doi:10.1111/joim.13640
150. Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40:920-938. doi:10.1016/j.ccell.2022.08.011
151. Deng S, Li C, Cao J, Cui Z, Du J, Fu Z, et al. Organ-on-a-chip meets artificial intelligence in drug evaluation. Theranostics. 2023;13:4526-4558. doi:10.7150/thno.87266
152. Liu J, Wang Q, Le Y, Hu M, Li C, An N, et al. 3D-bioprinting for precision microtissue engineering: advances, applications, and prospects. Adv Healthc Mater. 2025;14:e2403781. doi:10.1002/adhm.202403781
153. Shukla P, Yeleswarapu S, Heinrich MA, Prakash J, Pati F. Mimicking tumor microenvironment by 3D bioprinting: 3D cancer modeling. Biofabrication. 2022;14. doi:10.1088/1758-5090/ac6d11
154. Lawlor KT, Vanslambrouck JM, Higgins JW, Chambon A, Bishard K, Arndt D, et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat Mater. 2021;20:260-271. doi:10.1038/s41563-020-00853-9
155. Shukla P, Bera AK, Ghosh A, Kiranmai G, Pati F. Assessment and process optimization of high throughput biofabrication of immunocompetent breast cancer model for drug screening applications. Biofabrication. 2024;16. doi:10.1088/1758-5090/ad586b
156. Subtil B, Iyer KK, Poel D, Bakkerus L, Gorris MAJ, Cuenca Escalona J, et al. Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front Immunol. 2023;14:1105244. doi:10.3389/fimmu.2023.1105244
157. Magré L, Verstegen MMA, Buschow S, van der Laan LJW, Peppelenbosch M, Desai J. Emerging organoid–immune co-culture models for cancer research: from oncoimmunology to personalized immunotherapies. J Immunother Cancer. 2023;11:6290. doi:10.1136/jitc-2022-006290
158. Yang R, Yu Y. Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 2023;562:216180. doi:10.1016/j.canlet.2023.216180
159. Saorin G, Caligiuri I, Rizzolio F. Microfluidic organoids-on-a-chip: the future of human models. Semin Cell Dev Biol.2023;144:41-54. doi:10.1016/j.semcdb.2022.10.001
160. Lee JW, Hur J, Kwon YW, Chae CW, Choi JI, Hwang I, et al. KAI1 (CD82) is a key molecule to control angiogenesis and switch angiogenic milieu to quiescent state. J Hematol Oncol. 2021;14:148. doi:10.1186/s13045-021-01147-6
161. Luo Q, Wang J, Zhao W, Peng Z, Liu X, Li B, et al. Vasculogenic mimicry in carcinogenesis and clinical applications. J Hematol Oncol. 2020;13:19. doi:10.1186/s13045-020-00858-6
162. Quintard C, Tubbs E, Jonsson G, Jiao J, Wang J, Werschler N, et al. A microfluidic platform integrating functional vascularized organoids-on-chip. Nat Commun. 2024;15:1452. doi:10.1038/s41467-024-45710-4
163. Du Y, Wang YR, Bao QY, Xu XX, Xu C, Wang S, et al. Personalized vascularized tumor organoid-on-a-chip for tumor metastasis and therapeutic targeting assessment. Adv Mater. 2025;37:e2412815. doi:10.1002/adma.202412815
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Ling Wang, Qin Tang, Lin Zhong, Misi He, Xueping Zhu, Qian Zheng, Ling Long, Ya Wang, Qingxiu Jiang, Haixia Wang, Dongling Zou

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright
Authors publishing in Cancer Biome and Targeted Therapy retain full copyright of their work. By submitting a manuscript, authors grant the publisher (GCINC Press) a non-exclusive license to publish, distribute, and archive the article, and to identify itself as the original publisher.
License
All articles are published open access under the terms of the Creative Commons Attribution 4.0 International License (CC BY 4.0).
https://creativecommons.org/licenses/by/4.0/. This license permits unrestricted use, distribution, reproduction, and adaptation in any medium, including for commercial purposes, provided that:
- Proper attribution is given to the original author(s) and source,
- A link to the license is provided, and
- Any changes made are clearly indicated.
Author Rights
Authors retain the right to:
- Use their article in future works (e.g., books, theses, lectures)
- Share and archive the final published version on institutional repositories or personal websites
- Adapt or translate their work, or authorize others to do so, with proper citation
Reuse by Third Parties
Content is licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0). Third parties may copy, redistribute, remix, transform, and build upon the material for any purpose, including commercial use, provided that appropriate credit is given to the original author(s).
Archiving and Preservation
All articles are made freely available immediately upon publication, without embargo. Cancer Biome and Targeted Therapy is hosted on the Open Journal Systems (OJS) platform, developed by the Public Knowledge Project (PKP). The journal participates in long-term digital preservation through the PKP Preservation Network (PKP PN) using the LOCKSS system. Authors are encouraged to self-archive in institutional repositories, disciplinary archives, and preprint servers in accordance with the license terms.



