The Role of the Microbiome in Cancer Development
Keywords:
cancer microbiome, Tumor microenvironment, microbial dysbiosis, Drug resistance, Microbiota, Biofilms, Host-microbe interactionsAbstract
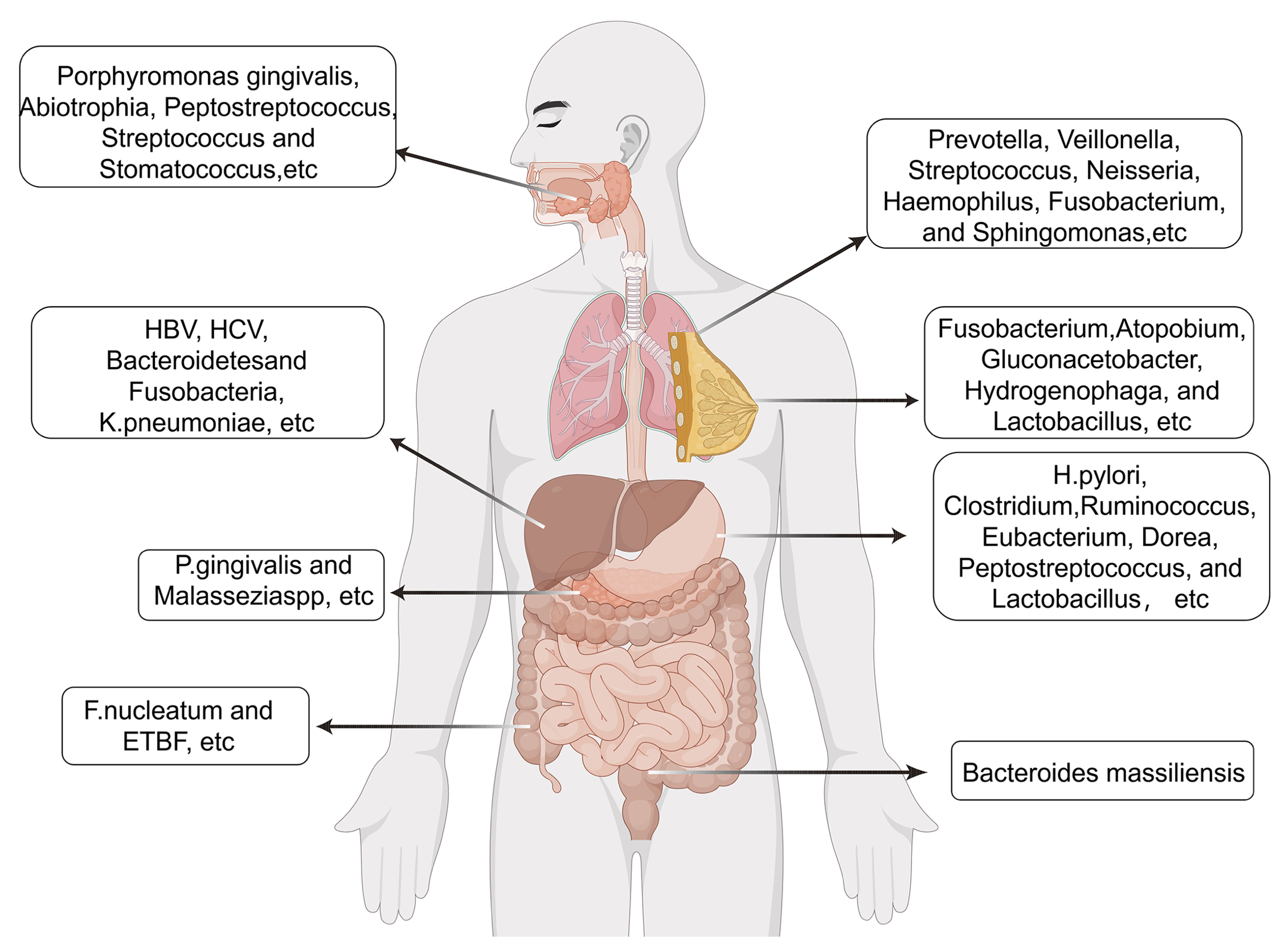
An estimated 38 trillion microbiome including bacteria, fungi, and viruses, reside in the human body, a population comparable in scale to the total number of somatic and germline cells. Microbial communities traditionally thought to be restricted to exposed anatomical sites such as the gastrointestinal tract, skin, oral cavity, and vaginal canal are now recognized to inhabit internal organs previously considered sterile. Technological advancements in sequencing and microbial detection have revealed the presence of low-biomass microbiota within various organs, including the lungs, mammary glands, liver, pancreas, prostate, and kidneys. The human microbiome constitutes an indispensable component of physiological processes, with the majority of microbial species exerting commensal or beneficial effects that support host health. Nevertheless, a subset of these microorganisms has been identified as pathogenic, with some demonstrating clear associations with cancer. This evolving understanding has given rise to the emerging concept of the cancer microbiome.
References
1. DiMaio D, Emu B, Goodman AL, Mothes W, Justice A. Cancer microbiology. J Natl Cancer Inst. 2022;114(5):651-663. doi:10.1093/jnci/djab212
2. Wang Y, Zhang R, Pu Y, Wang D, Wang Y, Wu X, et al. Sample collection, DNA extraction, and library construction protocols of the human microbiome studies in the International Human Phenome Project. Phenomics. 2023;3(3):300-308. doi:10.1007/s43657-023-00097-y
3. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5(10):3111-3122. PMID: 26609491. PMCID: PMC4656734
4. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer compared with benign mass-like lesions. Lung Cancer. 2016;102:89-95. doi:10.1016/j.lungcan.2016.10.016
5. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198(9):1188-1198. doi:10.1164/rccm.201710-2118OC
6. Chu S, Cheng Z, Yin Z, Xu J, Wu F, Jin Y, et al. Airway Fusobacterium is associated with poor response to immunotherapy in lung cancer. Onco Targets Ther. 2022;15:201-213. doi:10.2147/OTT.S348382
7. Derosa L, Routy B, Thomas AM, Iebba V, Zalcman G, Friard S, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med. 2022;28(2):315-324. doi:10.1038/s41591-021-01655-5
8. Teng L, Wang K, Chen W, Wang YS, Bi L. HYR-2 plays an anti-lung cancer role by regulating PD-L1 and Akkermansia muciniphila. Pharmacol Res. 2020;160:105086. doi:10.1016/j.phrs.2020.105086
9. Zhang Z, Liu D, Liu S, Zhang S, Pan Y. The role of Porphyromonas gingivalis outer membrane vesicles in periodontal disease and related systemic diseases. Front Cell Infect Microbiol. 2020;10:585917. doi:10.3389/fcimb.2020.585917
10. Pignatelli P, Nuccio F, Piattelli A, Curia MC. The role of Fusobacterium nucleatum in oral and colorectal carcinogenesis. Microorganisms. 2023;11(9):2358. doi:10.3390/microorganisms11092358
11. Bakhti SZ, Latifi-Navid S. Interplay and cooperation of Helicobacter pylori and gut microbiota in gastric carcinogenesis. BMC Microbiol. 2021;21(1):258. doi:10.1186/s12866-021-02315-x
12. Alipour M. Molecular mechanism of Helicobacter pylori-induced gastric cancer. J Gastrointest Cancer. 2021;52(1):23-30. doi:10.1007/s12029-020-00518-5
13. Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, et al. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. 2021;11(10):4945-4956. doi:10.7150/thno.55209
14. Rocken C. Predictive biomarkers in gastric cancer. J Cancer Res Clin Oncol. 2023;149(1):467-481. doi:10.1007/s00432-022-04408-0
15. Fletcher AA, Kelly MS, Eckhoff AM, Allen PJ. Revisiting the intrinsic mycobiome in pancreatic cancer. Nature. 2023;620(7972):E1-E6. doi:10.1038/s41586-023-06292-1
16. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. 2022;14(1):2073785. doi:10.1080/19490976.2022.2073785
17. Hayashi M, Ikenaga N, Nakata K, Luo H, Zhong P, Date S, et al. Intratumor Fusobacterium nucleatum promotes the progression of pancreatic cancer via the CXCL1–CXCR2 axis. Cancer Sci. 2023;114(9):3666-3678. doi:10.1111/cas.15901
18. Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023;31(2):159-172. doi:10.1016/j.tim.2022.08.010
19. Jasemi S, Molicotti P, Fais M, Cossu I, Simula ER, Sechi LA. Biological mechanisms of enterotoxigenic Bacteroides fragilis toxin: linking inflammation, colorectal cancer, and clinical implications. Toxins (Basel). 2025;17(6):305. doi:10.3390/toxins17060305
20. Chen B, Ramazzotti D, Heide T, Spiteri I, Fernandez-Mateos J, James C, et al. Contribution of pks(+) E. coli mutations to colorectal carcinogenesis. Nat Commun. 2023;14(1):7827. doi:10.1038/s41467-023-43329-5
21. Huang C, Mei S, Zhang X, Tian X. Inflammatory milieu related to dysbiotic gut microbiota promotes tumorigenesis of hepatocellular carcinoma. J Clin Gastroenterol. 2023;57(8):782-788. doi:10.1097/MCG.0000000000001883
22. Shi Q, Wang J, Zhou M, Zheng R, Zhang X, Liu B. Gut Lactobacillus contributes to the progression of breast cancer by affecting the anti-tumor activities of immune cells in the TME of tumor-bearing mice. Int Immunopharmacol. 2023;124(Pt B):111039. doi:10.1016/j.intimp.2023.111039
23. Peng F, Hu M, Su Z, Hu L, Guo L, Yang K. Intratumoral microbiota as a target for advanced cancer therapeutics. Adv Mater. 2024;36(38):2405331. doi:10.1002/adma.202405331
24. Barrett M, Hand CK, Shanahan F, Murphy T, O’Toole PW. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer. 2020;6(4):277-287. doi:10.1016/j.trecan.2020.01.019
25. Jiang M, Yang Z, Dai J, Wu T, Jiao Z, Yu Y, et al. Intratumor microbiome: selective colonization in the tumor microenvironment and a vital regulator of tumor biology. MedComm (2020). 2023;4(5):e376. doi:10.1002/mco2.376
26. Joo JE, Chu YL, Georgeson P, Walker R, Mahmood K, Clendenning M, et al. Intratumoral presence of the genotoxic gut bacteria pks(+) E. coli, enterotoxigenic Bacteroides fragilis, and Fusobacterium nucleatum and their association with clinicopathological and molecular features of colorectal cancer. Br J Cancer. 2024;130(5):728-740. doi:10.1038/s41416-023-02554-x
27. Wong CC, Yu J. pks(+) E. coli adhesins—the fine line between good and evil. Cell Host Microbe. 2025;33(1):1-3. doi:10.1016/j.chom.2024.12.007
28. Cao Y, Oh J, Xue M, Huh WJ, Wang J, González-Hernández JA, et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science. 2022;378(6618):eabm3233. doi:10.1126/science.abm3233
29. Khan S. Potential role of Escherichia coli DNA mismatch-repair proteins in colon cancer. Crit Rev Oncol Hematol. 2015;96(3):475-482. doi:10.1016/j.critrevonc.2015.05.002
30. Santos JC, Brianti MT, Almeida VR, Ortega MM, Fischer W, Haas R, et al. Helicobacter pylori infection modulates the expression of miRNAs associated with the DNA mismatch repair pathway. Mol Carcinog. 2017;56(4):1372-1379. doi:10.1002/mc.22590
31. Khan FH, Dervan E, Bhattacharyya DD, McAuliffe JD, Miranda KM, Glynn SA. The role of nitric oxide in cancer: master regulator or NOt? Int J Mol Sci. 2020;21(24):9393. doi:10.3390/ijms21249393
32. Zhao Y, Ye X, Xiong Z, Ihsan A, Ares I, Martínez M, et al. Cancer metabolism: the role of ROS in DNA damage and induction of apoptosis in cancer cells. Metabolites. 2023;13(7):796. doi:10.3390/metabo13070796
33. DeCaprio JA. Molecular pathogenesis of Merkel cell carcinoma. Annu Rev Pathol. 2021;16:69-91. doi:10.1146/annurev-pathmechdis-012419-032817
34. Torre-Castro J, Rodríguez M, Alonso-Alonso R, Mendoza Cembranos MD, Díaz-Alejo JF, Rebollo-González M, et al. LT and SOX9 expression are associated with gene sets that distinguish Merkel cell polyomavirus (MCPyV)-positive and MCPyV-negative Merkel cell carcinoma. Br J Dermatol. 2024;190(6):876-884. doi:10.1093/bjd/ljae033
35. Fernandes Q, Inchakalody VP, Bedhiafi T, Mestiri S, Taib N, Uddin S, et al. Chronic inflammation and cancer; the two sides of a coin. Life Sci. 2024;338:122390. doi:10.1016/j.lfs.2023.122390
36. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:1533033819867354. doi:10.1177/1533033819867354
37. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254-258. doi:10.1038/nature11465
38. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7-34. doi:10.3322/caac.21551
39. Tang Z, Liang D, Deubler EL, Sarnat JA, Chow SS, Diver WR, et al. Lung cancer metabolomics: a pooled analysis in the Cancer Prevention Studies. BMC Med. 2024;22(1):262. doi:10.1186/s12916-024-03473-1
40. Rodríguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Treat Res. 2016;170:25-46. doi:10.1007/978-3-319-40389-2_2
41. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018;24(13):3528-3538. doi:10.1016/j.celrep.2018.08.090
42. Kaderbhai C, Richard C, Fumet JD, Aarnink A, Foucher P, Coudert B, et al. Antibiotic use does not appear to influence response to nivolumab. Anticancer Res. 2017;37(6):3195-3200. doi:10.21873/anticanres.11680
43. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957-963. doi:10.1164/rccm.201104-0655OC
44. Pilette C, Ouadrhiri Y, Godding V, Vaerman JP, Sibille Y. Lung mucosal immunity: immunoglobulin-A revisited. Eur Respir J. 2001;18(3):571-588. doi:10.1183/09031936.01.00228801
45. Toma I, Siegel MO, Keiser J, Yakovleva A, Kim A, Davenport L, et al. Single-molecule long-read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol. 2014;52(11):3913-3921. doi:10.1128/JCM.01678-14
46. McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, et al. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365(17):1567-1575. doi:10.1056/NEJMoa1106955
47. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2):e16384. doi:10.1371/journal.pone.0016384
48. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi:10.1371/journal.pone.0008578
49. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep. 2019;9(1):12838. doi:10.1038/s41598-019-49195-w
50. Chung KF. Airway microbial dysbiosis in asthmatic patients: a target for prevention and treatment? J Allergy Clin Immunol. 2017;139(4):1071-1081. doi:10.1016/j.jaci.2017.02.004
51. Hewitt RJ, Lloyd CM. Regulation of immune responses by the airway epithelial cell landscape. Nat Rev Immunol. 2021;21(6):347-362. doi:10.1038/s41577-020-00477-9
52. Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, et al. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int J Biol Sci. 2019;15(11):2381-2392. doi:10.7150/ijbs.35980
53. Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. 2019;9:112. doi:10.3389/fcimb.2019.00112
54. Zhang WQ, Zhao SK, Luo JW, Dong XP, Hao YT, Li H, et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res. 2018;10(10):3171-3185. https://pmc.ncbi.nlm.nih.gov/articles/PMC6220220/
55. Wang H, Hu J, Ma Y, Abulimiti Y, Zhou Y. Lung commensal bacteria promote lung cancer progression through NK cell-mediated immunosuppressive microenvironment. Int J Med Sci. 2025;22(5):1039-1051. doi:10.7150/ijms.107026
56. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R, et al. Specific gut microbiome signature predicts early-stage lung cancer. Gut Microbes. 2020;11(4):1030-1042. doi:10.1080/19490976.2020.1737487
57. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6(2):471-474. doi:10.1038/ismej.2011.104
58. Qiao D, Wang Z, Lu Y, Wen X, Li H, Zhao H. A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am J Cancer Res. 2014;5(1):423-432. https://pmc.ncbi.nlm.nih.gov/articles/PMC4300720/
59. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17(1):163. doi:10.1186/s13059-016-1021-1
60. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957-963. doi:10.1164/rccm.201104-0655OC
61. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi:10.1371/journal.pone.0008578
62. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821-830. doi:10.1513/AnnalsATS.201501-029OC
63. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6(2):471-474. doi:10.1038/ismej.2011.104
64. Cameron SJ, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One. 2017;12(5):e0177062. doi:10.1371/journal.pone.0177062
65. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821-830. doi:10.1513/AnnalsATS.201501-029OC
66. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi:10.1038/nmicrobiol.2016.31
67. Oriano M, Gramegna A, Terranova L, Sotgiu G, Sulaiman I, Ruggiero L, et al. Sputum neutrophil elastase associates with microbiota and Pseudomonas aeruginosa in bronchiectasis. Eur Respir J. 2020;56(4):2000769. doi:10.1183/13993003.00769-2020
68. Tsay JC, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198(9):1188-1198. doi:10.1164/rccm.201710-2118OC
69. Mac Aogáin M, Dicker AJ, Mertsch P, Chotirmall SH. Infection and the microbiome in bronchiectasis. Eur Respir Rev. 2024;33(173):240038. doi:10.1183/16000617.0038-2024
70. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998-1013.e16. doi:10.1016/j.cell.2018.12.040
71. Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488-500. doi:10.1007/s13238-018-0548-1
72. Yamashita Y, Takeshita T. The oral microbiome and human health. J Oral Sci. 2017;59(2):201-206. doi:10.2334/josnusd.16-0856
73. Santacroce L, Passarelli PC, Azzolino D, Bottalico L, Charitos IA, Cazzolla AP, Colella M, Topi S, Garcia-Godoy F, D’Addona A. Oral microbiota in human health and disease: a perspective. Exp Biol Med (Maywood). 2023;248(15):1288-1301. doi:10.1177/15353702231187645
74. Kilian M. The oral microbiome—friend or foe? Eur J Oral Sci. 2018;126(Suppl 1):5-12. doi:10.1111/eos.12527
75. Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777-6787. doi:10.1158/0008-5472.CAN-17-1296
76. Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95(7):569-574. doi:10.1111/j.1349-7006.2004.tb02488.x
77. Snider EJ, Compres G, Freedberg DE, Giddins MJ, Khiabanian H, Lightdale CJ, et al. Barrett’s esophagus is associated with a distinct oral microbiome. Clin Transl Gastroenterol. 2018;9(3):e135. doi:10.1038/s41424-018-0005-8
78. Vogtmann E, Han Y, Caporaso JG, Bokulich N, Mohamadkhani A, Moayyedkazemi A, et al. Oral microbial community composition is associated with pancreatic cancer: a case-control study in Iran. Cancer Med. 2020;9(2):797-806. doi:10.1002/cam4.2660
79. Del Castillo E, Meier R, Chung M, Koestler DC, Chen T, Paster BJ, et al. The microbiomes of pancreatic and duodenum tissue overlap and are highly subject-specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol Biomarkers Prev. 2019;28(2):370-383. doi:10.1158/1055-9965.EPI-18-0542
80. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454-1463. doi:10.1136/gutjnl-2017-314814
81. Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122-128. doi:10.4103/jomfp.JOMFP_304_18
82. Demmitt BA, Corley RP, Huibregtse BM, Keller MC, Hewitt JK, McQueen MB, et al. Genetic influences on the human oral microbiome. BMC Genomics. 2017;18(1):659. doi:10.1186/s12864-017-4008-8
83. Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162(2):22-38. doi:10.1016/j.imlet.2014.08.017
84. Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 2018;3(6):e00187-18. doi:10.1128/mSystems.00187-18
85. Oren A, Garrity GM. Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. 2021;71(10):005056. doi:10.1099/ijsem.0.005056
86. McLean JS. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:98. doi:10.3389/fcimb.2014.00098
87. Baker JL, Mark Welch JL, Kauffman KM, McLean JS, He X. The oral microbiome: diversity, biogeography and human health. Nat Rev Microbiol. 2024;22(2):89-104. doi:10.1038/s41579-023-00963-6
88. Diaz P, Dongari-Bagtzoglou A. Critically appraising the significance of the oral mycobiome. J Dent Res. 2021;100(2):133-140. doi:10.1177/0022034520956975
89. Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. doi:10.1186/1471-2164-14-623
90. Turner SA, Butler G. The Candida pathogenic species complex. Cold Spring Harb Perspect Med. 2014;4(9):a019778. doi:10.1101/cshperspect.a019778
91. Ahmad KM, Kokošar J, Guo X, Gu Z, Ishchuk OP, Piskur J. Genome structure and dynamics of the yeast pathogen Candida glabrata. FEMS Yeast Res. 2014;14(4):529-535. doi:10.1111/1567-1364.12145
92. Hong BY, Hoare A, Cardenas A, Dupuy A, Choquette L, Salner A, et al. The salivary mycobiome contains 2 ecologically distinct mycotypes. J Dent Res. 2020;99(6):730-738. doi:10.1177/0022034520915879
93. Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9(3):e90899. doi:10.1371/journal.pone.0090899
94. Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10(3):e1003933. doi:10.1371/journal.ppat.1003933
95. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi:10.3322/caac.21660
96. Irfan M, Delgado RZR, Frias-Lopez J. The oral microbiome and cancer. Front Immunol. 2020;11:591088. doi:10.3389/fimmu.2020.591088
97. Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2020;9:476. doi:10.3389/fcimb.2019.00476
98. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3(4):209-215. doi:10.4248/ijos11075
99. Sayehmiri F, Sayehmiri K, Asadollahi K, Soroush S, Bogdanovic L, Jalilian FA, et al. The prevalence rate of Porphyromonas gingivalis and its association with cancer: a systematic review and meta-analysis. Int J Immunopathol Pharmacol. 2015;28(2):160-167. doi:10.1177/0394632015586144
100. Wen L, Mu W, Lu H, Wang X, Fang J, Jia Y, et al. Porphyromonas gingivalis promotes oral squamous cell carcinoma progression in an immune microenvironment. J Dent Res. 2020;99(6):666-675. doi:10.1177/0022034520909312
101. Rai AK, Panda M, Das AK, Rahman T, Das R, Das K, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. 2021;203(1):137-152. doi:10.1007/s00203-020-02011-w
102. Mammen MJ, Scannapieco FA, Sethi S. Oral–lung microbiome interactions in lung diseases. Periodontol 2000. 2020;83(1):234-241. doi:10.1111/prd.12301
103. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59-64. doi:10.1038/nature13568
104. Acharya C, Sahingur SE, Bajaj JS. Microbiota, cirrhosis, and the emerging oral–gut–liver axis. JCI Insight. 2017;2(19):e94416. doi:10.1172/jci.insight.94416
105. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359-365. doi:10.1126/science.aan4526
106. Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182(2):447-462.e14. doi:10.1016/j.cell.2020.05.048
107. Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215-225. doi:10.1016/j.chom.2016.07.006
108. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case–control study. Gut. 2018;67(1):120-127. doi:10.1136/gutjnl-2016-312580
109. Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjonneland A, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62(12):1764-1770. doi:10.1136/gutjnl-2012-303006
110. Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. doi:10.7717/peerj.1373
111. Karpiński TM. The microbiota and pancreatic cancer. Gastroenterol Clin North Am. 2019;48(3):447-464. doi:10.1016/j.gtc.2019.04.008
112. Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209-7220. doi:10.18632/oncotarget.3109
113. Wei AL, Li M, Li GQ, Wang X, Hu WM, Li ZL, et al. Oral microbiome and pancreatic cancer. World J Gastroenterol. 2020;26(48):7679-7692. doi:10.3748/wjg.v26.i48.7679
114. Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582-588. doi:10.1136/gutjnl-2011-300784
115. Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5(12):1749-1768. doi:doi:10.1001/jamaoncol.2019.2996
116. Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23(4):713-739. doi:10.1128/CMR.00011-10
117. Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, et al. Clinical significance of four molecular subtypes of gastric cancer identified by The Cancer Genome Atlas Project. Clin Cancer Res. 2017;23(15):4441-4449. doi:10.1158/1078-0432.CCR-16-2211
118. Mommersteeg MC, Yu J, Peppelenbosch MP, Fuhler GM. Genetic host factors in Helicobacter pylori-induced carcinogenesis: emerging new paradigms. Biochim Biophys Acta Rev Cancer. 2018;1869(1):42-52. doi:10.1016/j.bbcan.2017.11.003
119. Castano-Rodríguez N, Kaakoush NO, Mitchell HM. Pattern-recognition receptors and gastric cancer. Front Immunol. 2014;5:336. doi:10.3389/fimmu.2014.00336
120. Moss SF. The clinical evidence linking Helicobacter pylori to gastric cancer. Cell Mol Gastroenterol Hepatol. 2017;3(2):183-191. doi:10.1016/j.jcmgh.2016.12.001
121. Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720-741. doi:10.1128/CMR.10.4.720
122. Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type gastric cancer. Sci Rep. 2014;4:4202. doi:10.1038/srep04202
123. Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407-416. doi:10.1111/hel.12145
124. Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7(1):15957. doi:10.1038/s41598-017-16289-2
125. Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7(3):e33418. doi:10.1371/journal.pone.0033418
126. Serban DE. Gastrointestinal cancers: influence of gut microbiota, probiotics and prebiotics. Cancer Lett. 2014;345(2):258-270. doi:10.1016/j.canlet.2013.08.013
127. Kidd M, Modlin IM. A century of Helicobacter pylori: paradigms lost–paradigms regained. Digestion. 1998;59(1):1-15. doi:10.1159/000007461
128. Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;321(8336):1273-1275. doi:10.1016/S0140-6736(83)92719-8
129. Llorca L, Pérez-Pérez G, Urruzuno P, Martinez MJ, Iizumi T, Gao Z, et al. Characterization of the gastric microbiota in a pediatric population according to Helicobacter pylori status. Pediatr Infect Dis J. 2017;36(2):173-178. doi:10.1097/INF.0000000000001383
130. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688-693. doi:10.1038/sj.embor.7400731
131. Minaga K, Watanabe T, Kamata K, Asano N, Kudo M. Nucleotide-binding oligomerization domain 1 and Helicobacter pylori infection: a review. World J Gastroenterol. 2018;24(16):1725-1733. doi:10.3748/wjg.v24.i16.1725
132. Yang J, Zhou X, Liu X, Ling Z, Ji F. Role of the gastric microbiome in gastric cancer: from carcinogenesis to treatment. Front Microbiol. 2021;12:641322. doi:10.3389/fmicb.2021.641322
133. Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, et al. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9(1):19. doi:10.1038/s41572-023-00431-8
134. Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol. 2017;112(2):212-239. doi:10.1038/ajg.2016.563
135. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859-904. doi:10.1152/physrev.00045.2009
136. Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer J. 2014;20(3):211-216. doi:10.1097/PPO.0000000000000043
137. Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863-1873. doi:10.1053/j.gastro.2009.01.073
138. Szabò I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, et al. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18(20):5517-5527. doi:10.1093/emboj/18.20.5517
139. Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, et al. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. J Pathol. 2007;213(2):140-151. doi:10.1002/path.2218
140. Bimela JS, Nanfack AJ, Yang P, Dai S, Kong XP, Torimiro JN, et al. Antiretroviral imprints and genomic plasticity of HIV-1 pol in non-clade B: implications for treatment. Front Microbiol. 2022;12:812391. doi:10.3389/fmicb.2021.812391
141. Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185(20):3807-3822.e12. doi:10.1016/j.cell.2022.09.015
142. Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X, et al. Integrative identification of Epstein–Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology. 2014;147(6):1350-1362.e4. doi:10.1053/j.gastro.2014.08.036
143. Yang J, Liu Z, Zeng B, Hu G, Gan R. Epstein–Barr virus-associated gastric cancer: a distinct subtype. Cancer Lett. 2020;495:191-199. doi:10.1016/j.canlet.2020.09.019
144. Sun K, Jia K, Lv H, Wang SQ, Wu Y, Lei H, et al. EBV-positive gastric cancer: current knowledge and future perspectives. Front Oncol. 2020;10:583463. doi:10.3389/fonc.2020.583463
145. Niedźwiedzka-Rystwej P, Grywalska E, Hrynkiewicz R, Wolacewicz M, Becht R, Roliński J. The double-edged sword role of viruses in gastric cancer. Cancers (Basel). 2020;12(6):1680. doi:10.3390/cancers12061680
146. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33. doi:10.3322/caac.21654
147. Ro C, Chai W, Yu VE, Yu R. Pancreatic neuroendocrine tumors: biology, diagnosis, and treatment. Chin J Cancer. 2013;32(6):312-324. doi:10.5732/cjc.012.10295
148. Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363(9414):1049-1057. doi:10.1016/S0140-6736(04)15841-8149. Cruz MS, Tintelnot J, Gagliani N. Roles of microbiota in pancreatic cancer development and treatment. Gut Microbes. 2024;16(1):2320280. doi:10.1080/19490976.2024.2320280
150. Kaźmierczak-Siedlecka K, Dvorák A, Folwarski M, Daca A, Przewlócka K, Makarewicz W. Fungal gut microbiota dysbiosis and its role in colorectal, oral, and pancreatic carcinogenesis. Cancers (Basel). 2020;12(5):1326. doi:10.3390/cancers12051326
151. Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18(1):97. doi:10.1186/s12943-019-1008-0
152. Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2015;12(2):77-87. doi:10.1038/nrgastro.2014.188
153. Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8(6):e66019. doi:10.1371/journal.pone.0066019
154. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264-267. doi:10.1038/s41586-019-1608-2
155. Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep. 2015;5:7980. doi:10.1038/srep07980
156. Li S, Fuhler GM, Bn N, Jose T, Bruno MJ, Peppelenbosch MP, et al. Pancreatic cyst fluid harbors a unique microbiome. Microbiome. 2017;5(1):147. doi:10.1186/s40168-017-0363-6
157. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403-416. doi:10.1158/2159-8290.CD-17-1134
158. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512-516. doi:10.1038/nature24462
159. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973-980. doi:10.1126/science.aay9189
160. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795-806.e12. doi:10.1016/j.cell.2019.07.008
161. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156-1160. doi:10.1126/science.aah5043
162. Fiorillo L, Cervino G, Laino L, D’Amico C, Mauceri R, Tozum TF, et al. Porphyromonas gingivalis, periodontal and systemic implications: a systematic review. Dent J (Basel). 2019;7(4):114. doi:10.3390/dj7040114
163. Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels, and orodigestive cancer mortality. Carcinogenesis. 2012;33(5):1055-1058. doi:10.1093/carcin/bgs112
164. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case–control study. Gut. 2018;67(1):120-127. doi:10.1136/gutjnl-2016-312580
165. Huang J, Roosaar A, Axéll T, Ye W. A prospective cohort study on poor oral hygiene and pancreatic cancer risk. Int J Cancer. 2016;138(2):340-347. doi:10.1002/ijc.29710
166. Jin Y, Gao H, Chen H, Wang J, Chen M, Li G, et al. Identification and impact of hepatitis B virus DNA and antigens in pancreatic cancer tissues and adjacent non-cancerous tissues. Cancer Lett. 2013;335(2):447-454. doi:10.1016/j.canlet.2013.03.001
167. Shimoda T, Shikata T, Karasawa T, Tsukagoshi S, Yoshimura M, Sakurai I. Light microscopic localization of hepatitis B virus antigens in the human pancreas: possibility of multiplication of hepatitis B virus in the human pancreas. Gastroenterology. 1981;81(6):998-1005. doi:10.1016/S0016-5085(81)80004-2
168. Arafa A, Eshak ES, Abdel Rahman TA, Anwar MM. Hepatitis C virus infection and risk of pancreatic cancer: a meta-analysis. Cancer Epidemiol. 2020;65:101691. doi:10.1016/j.canep.2020.101691
169. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7-33. doi:10.3322/caac.21708
170. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322-340. doi:10.1053/j.gastro.2019.06.048
171. Wang J, Chen W-D, Wang Y-D. The relationship between gut microbiota and inflammatory diseases: the role of macrophages. Front Microbiol. 2020;11:1065. doi:10.3389/fmicb.2020.01065
172. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223-237. doi:10.1038/s41575-019-0258-z
173. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299-306. doi:10.1101/gr.126516.111
174. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207-215. doi:10.1016/j.chom.2013.07.007
175. Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res (Phila). 2017;10(7):398-409. doi:10.1158/1940-6207.CAPR-16-0178
176. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195-206. doi:10.1016/j.chom.2013.07.012
177. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590. doi:10.1038/s41598-017-11237-6
178. Guinney J, Dienstmann R, Wang X, De Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350-1356. doi:10.1038/nm.3967
179. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443-1448. doi:10.1126/science.aal5240
180. Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22(2):349-369. doi:10.1128/CMR.00053-08
181. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, Shields CED, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018; 23(2):203-214.e5. doi:10.1016/j.chom.2018.01.007
182. Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016-1022. doi:10.1038/nm.2015
183. Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe JL, et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5(10):1098-1109. doi:10.1158/2159-8290.CD-15-0447
184. DeDecker L, Coppedge B, Avelar-Barragan J, Karnes W, Whiteson K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes. 2021;13(1):1-12. doi:10.1080/19490976.2020.1854641
185. Goodwin AC, Shields CED, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354-15359. doi:10.1073/pnas.1010203108
186. Allen J, Hao S, Sears CL, Timp W. Epigenetic changes induced by Bacteroides fragilis toxin. Infect Immun. 2019;87(6):e00447-18. doi:10.1128/IAI.00447-18
187. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289-300. doi:10.1136/gutjnl-2018-317200
188. Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848-851. doi:10.1126/science.1127059
189. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3(12):1683-1691. doi:10.1001/jamaoncol.2017.3055
190. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589-604. doi:10.1038/s41575-019-0186-y
191. Thilakarathna W, Rupasinghe HPV, Ridgway ND. Mechanisms by which probiotic bacteria attenuate the risk of hepatocellular carcinoma. Int J Mol Sci. 2021;22(5):2606. doi:10.3390/ijms22052606
192. Zhang N, Gou Y, Liang S, Chen N, Liu Y, He Q, et al. Dysbiosis of gut microbiota promotes hepatocellular carcinoma progression by regulating the immune response. J Immunol Res. 2021;2021:4973589. doi:10.1155/2021/4973589
193. Zheng R, Wang G, Pang Z, Ran N, Gu Y, Guan X, et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020;9(12):4232-4250. doi:10.1002/cam4.3045
194. Wang X, Fang Y, Liang W, Cai Y, Wong CC, Wang J, et al. Gut-liver translocation of pathogen Klebsiella pneumoniae promotes hepatocellular carcinoma in mice. Nat Microbiol. 2025;10(1):169-184. doi:10.1038/s41564-024-01890-9
195. Song Y, Lau HC, Zhang X, Yu J. Bile acids, gut microbiota, and therapeutic insights in hepatocellular carcinoma. Cancer Biol Med. 2023;21(2):144-162. doi:10.20892/j.issn.2095-3941.2023.0394
196. Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes. 2014;5(4):441-445. doi:10.4161/gmic.29599
197. Shen S, Khatiwada S, Behary J, Kim R, Zekry A. Modulation of the gut microbiome to improve clinical outcomes in hepatocellular carcinoma. Cancers (Basel). 2022;14(9):2099. doi:10.3390/cancers14092099
198. Ram AK, Pottakat B, Vairappan B. Increased systemic zonula occludens1 associated with inflammation and independent biomarker in patients with hepatocellular carcinoma. BMC Cancer. 2018;18(1):572. doi:10.1186/s12885-018-4484-5
199. Guo S, Al-Sadi R, Said HM, Ma TY. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am J Pathol. 2013;182(2):375-387. doi:10.1016/j.ajpath.2012.10.014
200. Behary J, Raposo AE, Amorim NML, Zheng H, Gong L, McGovern E, et al. Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2⁻/⁻ mouse model. BMC Microbiol. 2021;21(1):113. doi:10.1186/s12866-021-02171-9
201. Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25(1):79. doi:10.1186/s12929-018-0483-8
202. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504-516. doi:10.1016/j.ccr.2012.02.007
203. Yu LX, Yan HX, Liu Q, Yang W, Wu HP, Dong W, et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52(4):1322-1333. doi:10.1002/hep.23845
204. Ranaee M, Torabi H, Azhganzad N, Shirini K, Hosseini AS, Hajian K. The relationship between tumor budding and patient survival in breast cancer. Clin Pathol. 2024;17:2632010X241235543. doi:10.1177/2632010X241235543
205. Wu J, Fan D, Shao Z, Xu B, Ren G, Jiang Z, et al. CACA guidelines for holistic integrative management of breast cancer. Holist Integr Oncol. 2022;1(1):7. doi:10.1007/s44178-022-00007-8
206. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12-49. doi:10.3322/caac.21820
207. Xu Y, Gong M, Wang Y, Yang Y, Liu S, Zeng Q. Global trends and forecasts of breast cancer incidence and deaths. Sci Data. 2023;10(1):334. doi:10.1038/s41597-023-02253-5
208. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263. doi:10.3322/caac.21834
209. Luo L, Chen Y, Ma Q, Huang Y, Xu L, Shu K, et al. Ginger volatile oil inhibits the growth of MDA-MB-231 in the bisphenol A environment by altering gut microbial diversity. Heliyon. 2024;10(2):e24388. doi:10.1016/j.heliyon.2024.e24388
210. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271-285. doi:10.1038/nrc.2017.13
211. Smith A, Pierre JF, Makowski L, Tolley E, Lyn-Cook B, Lu L, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of Non-Hispanic Black and Non-Hispanic White women. Sci Rep. 2019;9(1):11940. doi:10.1038/s41598-019-48348-1
212. Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS One. 2017;12(11):e0188873. doi:10.1371/journal.pone.0188873
213. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973-980. doi:10.1126/science.aay9189
214. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. 2016;6:30751. doi:10.1038/srep30751
215. Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9(1):e83744. doi:10.1371/journal.pone.0083744
216. Meng Z, Ye Z, Zhu P, Zhu J, Fang S, Qiu T, et al. New developments and opportunities of microbiota in treating breast cancers. Front Microbiol. 2022;13:818793. doi:10.3389/fmicb.2022.818793
217. Thu MS, Chotirosniramit K, Nopsopon T, Hirankarn N, Pongpirul K. Human gut, breast, and oral microbiome in breast cancer: a systematic review and meta-analysis. Front Oncol. 2023;13:1144021. doi:10.3389/fonc.2023.1144021
218. Parida S, Sharma D. The microbiome–estrogen connection and breast cancer risk. Cells. 2019;8(12):1642. doi:10.3390/cells8121642
219. Kwa M, Plottel CS, Blaser MJ, Adams S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst. 2016;108(8):djw029. doi:10.1093/jnci/djw029
220. Bernardo G, Le Noci V, Di Modica M, Montanari E, Triulzi T, Pupa SM, et al. The emerging role of the microbiota in breast cancer progression. Cells. 2023;12(15):1945. doi:10.3390/cells12151945
221. Virtanen V, Paunu K, Ahlskog JK, Varnai R, Sipeky C, Sundvall M. PARP inhibitors in prostate cancer—the preclinical rationale and current clinical development. Genes (Basel). 2019;10(8):565. doi:10.3390/genes10080565
222. Testa U, Castelli G, Pelosi E. Cellular and molecular mechanisms underlying prostate cancer development: therapeutic implications. Medicines (Basel). 2019;6(3):82. doi:10.3390/medicines6030082
223. Kopp W. How Western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab Syndr Obes. 2019;12:2221-2236. doi:10.2147/DMSO.S216791
224. Massari F, Mollica V, Di Nunno V, Gatto L, Santoni M, Scarpelli M, et al. The human microbiota and prostate cancer: friend or foe? Cancers (Basel). 2019;11(4):459. doi:10.3390/cancers11040459
225. Porter CM, Shrestha E, Peiffer LB, Sfanos KS. The microbiome in prostate inflammation and prostate cancer. Prostate Cancer Prostatic Dis. 2018;21(3):345-354. doi:10.1038/s41391-018-0041-1
226. Liss MA, White JR, Goros M, Gelfond J, Leach R, Johnson-Pais T, et al. Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur Urol. 2018;74(5):575-582. doi:10.1016/j.eururo.2018.06.033
227. Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology. 2018;111:122-128. doi:10.1016/j.urology.2017.08.039
228. Matsushita M, Fujita K, Hatano K, De Velasco MA, Uemura H, Nonomura N. Connecting the dots between the gut–IGF-1–prostate axis: a role of IGF-1 in prostate carcinogenesis. Front Endocrinol (Lausanne). 2022;13:852382. doi:10.3389/fendo.2022.852382
229. Fujita K, Nonomura N. Role of androgen receptor in prostate cancer: a review. World J Mens Health. 2019;37(3):288-295. doi:10.5534/wjmh.180040
230. Matsushita M, Fujita K, Motooka D, Hatano K, Hata J, Nishimoto M, et al. Firmicutes in gut microbiota correlate with blood testosterone levels in elderly men. World J Mens Health. 2022;40(3):517-525. doi:10.5534/wjmh.210190
231. Soheili M, Keyvani H, Soheili M, Nasseri S. Human papillomavirus: a review of epidemiology, carcinogenesis, diagnostic methods, and treatment of all HPV-related cancers. Med J Islam Repub Iran. 2021;35:65. doi:10.47176/mjiri.35.65
232. Smith JS, Melendy A, Rana RK, Pimenta JM. Age-specific prevalence of infection with human papillomavirus in females: a global review. J Adolesc Health. 2008;43(4 Suppl):S5-S25. doi:10.1016/j.jadohealth.2008.07.009
233. Watson M, Saraiya M, Benard V, Coughlin SS, Flowers L, Cokkinides V, et al. Burden of cervical cancer in the United States, 1998–2003. Cancer. 2008;113(10 Suppl):2855-2864. doi:10.1002/cncr.23756
234. Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210(11):1723-1733. doi:10.1093/infdis/jiu330
235. Laniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynecological cancer development, prevention, and therapy. Nat Rev Urol. 2020;17(4):232-250. doi:10.1038/s41585-020-0286-z
236. Makker V, MacKay H, Ray-Coquard I, Levine DA, Westin SN, Aoki D, et al. Endometrial cancer. Nat Rev Dis Primers. 2021;7(1):88. doi:10.1038/s41572-021-00324-8
237. Walther-António MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8(1):122. doi:10.1186/s13073-016-0368-y
238. Caselli E, Soffritti I, D’Accolti M, Piva I, Greco P, Bonaccorsi G. Atopobium vaginae and Porphyromonas somerae induce proinflammatory cytokine expression in endometrial cells: a possible implication for endometrial cancer? Cancer Manag Res. 2019;11:8571-8575. doi:10.2147/CMAR.S217362
239. Smolarz B, Biernacka K, Lukasiewicz H, Samulak D, Piekarska E, Romanowicz H, et al. Ovarian cancer - epidemiology, classification, pathogenesis, treatment, and estrogen receptor’ molecular backgrounds. Int J Mol Sci. 2025;26(10):4611. doi:10.3390/ijms26104611
240. Zhang M, Mo J, Huang W, Bao Y, Luo X, Yuan L. The ovarian cancer-associated microbiome contributes to the tumor’s inflammatory microenvironment. Front Cell Infect Microbiol. 2024;14:1440742. doi:10.3389/fcimb.2024.1440742
241. Wang Q, Zhao L, Han L, Fu G, Tuo X, Ma S, et al. Differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J Ovarian Res. 2020;13(1):8. doi:10.1186/s13048-019-0603-4
242. Yu B, Liu C, Proll SC, Manhardt E, Liang S, Srinivasan S, et al. Identification of fallopian tube microbiota and its association with ovarian cancer. eLife. 2024;12:RP89830. doi:10.7554/eLife.89830
243. Zhou B, Sun C, Huang J, Xia M, Guo E, Li N, et al. Biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. 2019;9(1):1691. doi:10.1038/s41598-018-38031-2
244. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, et al. The ovarian cancer oncobiome. Oncotarget. 2017;8(22):36225-36245. doi:10.18632/oncotarget.16717
245. Blanco JR, Del Campo R, Avendaño-Ortiz J, Laguna-Olmos M, Carnero A. The role of microbiota in ovarian cancer: implications for treatment response and therapeutic strategies. Cells. 2025;14(22):1813. doi:doi:10.3390/cells14221813
246. Huang Q, Wei X, Li W, Ma Y, Chen G, Zhao L, et al. Endogenous Propionibacterium acnes promotes ovarian cancer progression via regulation of the Hedgehog signaling pathway. Cancers (Basel). 2022;14(21):5178. doi:10.3390/cancers14215178
247. Kawahara N, Yamanaka S, Nishikawa K, Matsuoka M, Maehana T, Kawaguchi R, et al. Endogenous microbacteria can contribute to ovarian carcinogenesis by reducing iron concentration in cysts: a pilot study. Microorganisms. 2024;12(3):538. doi:10.3390/microorganisms12030538
248. McFarland LV. From yaks to yogurt: the history, development, and current use of probiotics. Clin Infect Dis. 2015;60(Suppl 2):S85-S90. doi:10.1093/cid/civ054
249. O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol. 2017;2:17057. doi:10.1038/nmicrobiol.2017.57
250. Shen H, Zhao Z, Zhao Z, Chen Y, Zhang L. Native and engineered probiotics: promising agents against related systemic and intestinal diseases. Int J Mol Sci. 2022;23(2):594. doi:10.3390/ijms23020594
251. Mahdavi M, Laforest-Lapointe I, Massé E. Preventing colorectal cancer through prebiotics. Microorganisms. 2021;9(6):1325. doi:10.3390/microorganisms9061325
252. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8(3):92. doi:10.3390/foods8030092
253. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. 2019;234(10):17127-17143. doi:10.1002/jcp.28473
254. Zhang M, Liu C, Tu J, Tang M, Ashrafizadeh M, Nabavi N, et al. Advances in cancer immunotherapy: historical perspectives, current developments, and future directions. Mol Cancer. 2025;24:136. doi:10.1186/s12943-025-02305-x
255. Hua D, Yang Q, Li X, Zhou X, Kang Y, Zhao Y, et al. The combination of Clostridium butyricum and Akkermansia muciniphila mitigates DSS-induced colitis and attenuates colitis-associated tumorigenesis by modulating gut microbiota and reducing CD8(+) T cells in mice. mSystems. 2025;10(2):e01567-24. doi:10.1128/msystems.01567-24
256. Saeed M, Shoaib A, Kandimalla R, Javed S, Almatroudi A, Gupta R, et al. Microbe-based therapies for colorectal cancer: advantages and limitations. Semin Cancer Biol. 2022;86(Pt 3):652-665. doi:10.1016/j.semcancer.2021.05.018
257. Gan BK, Rullah K, Yong CY, Ho KL, Omar AR, Alitheen NB, et al. Targeted delivery of 5-fluorouracil-1-acetic acid (5-FA) to cancer cells overexpressing epithelial growth factor receptor (EGFR) using virus-like nanoparticles. Sci Rep. 2020;10(1):16867. doi:10.1038/s41598-020-73967-4
258. Montassier E, Batard E, Massart S, Gastinne T, Carton T, Caillon J, et al. 16S rRNA gene pyrosequencing reveals a shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol. 2014;67(3):690-699. doi:10.1007/s00248-013-0355-4
259. van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, et al. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49(2):262-270. doi:10.1086/599346
260. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510-1519. doi:10.1136/gutjnl-2019-320204
261. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356-365. doi:10.1038/nrgastro.2017.20F
262. Schwan A, Sjölin S, Trottestam U, Aronsson B. Relapsing Clostridium difficile enterocolitis cured by rectal infusion of homologous faeces. Lancet. 1983;2(8354):845. doi:10.1016/S0140-6736(83)90753-5
263. Yang R, Chen Z, Cai J. Fecal microbiota transplantation: emerging applications in autoimmune diseases. J Autoimmun. 2023;141:103038. doi:10.1016/j.jaut.2023.103038
264. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13(9):508-516. doi:10.1038/nrgastro.2016.98
265. Chang CW, Lee HC, Li LH, Chiang Chiau JS, Wang TE, Chuang WH, et al. Fecal microbiota transplantation prevents intestinal injury, upregulation of Toll-like receptors, and 5-fluorouracil/oxaliplatin-induced toxicity in colorectal cancer. Int J Mol Sci. 2020;21(2):386. doi:10.3390/ijms21020386
266. Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, Iqbal TH. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther. 2017;46(5):479-493. doi:10.1111/apt.14201
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Yangyun Wang, Youyang Shi, Jingjing Duan, Haijia Tang, Sheng Liu, Jianfeng Yang

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright
Authors publishing in Cancer Biome and Targeted Therapy retain full copyright of their work. By submitting a manuscript, authors grant the publisher (GCINC Press) a non-exclusive license to publish, distribute, and archive the article, and to identify itself as the original publisher.
License
All articles are published open access under the terms of the Creative Commons Attribution 4.0 International License (CC BY 4.0).
https://creativecommons.org/licenses/by/4.0/. This license permits unrestricted use, distribution, reproduction, and adaptation in any medium, including for commercial purposes, provided that:
- Proper attribution is given to the original author(s) and source,
- A link to the license is provided, and
- Any changes made are clearly indicated.
Author Rights
Authors retain the right to:
- Use their article in future works (e.g., books, theses, lectures)
- Share and archive the final published version on institutional repositories or personal websites
- Adapt or translate their work, or authorize others to do so, with proper citation
Reuse by Third Parties
Content is licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0). Third parties may copy, redistribute, remix, transform, and build upon the material for any purpose, including commercial use, provided that appropriate credit is given to the original author(s).
Archiving and Preservation
All articles are made freely available immediately upon publication, without embargo. Cancer Biome and Targeted Therapy is hosted on the Open Journal Systems (OJS) platform, developed by the Public Knowledge Project (PKP). The journal participates in long-term digital preservation through the PKP Preservation Network (PKP PN) using the LOCKSS system. Authors are encouraged to self-archive in institutional repositories, disciplinary archives, and preprint servers in accordance with the license terms.



